Testing of dogs: Genetic profile DNA STR (ISAG2006)
Related tests
- DNA profile of the dog - combination of STR (ISAG2006) and SNP (ISAG2020)
- DSRA + DNA profile combination of DSRA and DNA profile tests for Cane Corso
Genetic profile
A genetic profile, sometimes referred to as a DNA profile, is a set of genetic markers that allow an individual to be uniquely identified. A genetic marker is a specific part of the genetic code of DNA that varies between individuals. A genetic profile is immutable throughout life and cannot be falsified or destroyed. It is used to identify an individual, verify parentage and relationships or to optimise breeding.
.
Genetic profiling possibilities
Genetic profiling serves as a reliable lifelong identification in many cases:
- In case of loss or theft of a dog - It allows to prove the identity of an individual in case a lost or stolen dog is found.
- In the event of a malfunctioning microchip - If the microchip the dog is carrying stops working, the dog's identity can be proven, and a new chipping can be performed.
- During insemination - Used to identify the semen
It is used to verify parentage:
- For routine pedigree checks or when parentage is in doubt - Used to verify reliably paternity or maternity after intentional or accidental mating or after insemination.
- Double mating - For some dog breeds it is possible to apply for approval of double mating. In this case it is necessary that the mother, all puppies and both fathers have DNA profiles and paternity of the puppies has been established.
- In the case of “clear by parents” verification - If both parents have been tested for a recessive genetic defect observed in a particular breed with negative results, i.e. both are healthy, their offspring can be designated as “clear by parents” when parentage is established by genetic profile. It is recommended to use it for every other generation only.
- If it is not possible to verify the parents, other family relationships can be verified - Sibling, grandson/grandfather, nephew/uncle... the more related individuals included in the test, the more accurate the result is obtained (the more accurate the probability of relationship)
In breeding it serves as a tool for:
- Selection of the optimal breeding pair - By comparing genetic profiles, the most suitable combinations of individuals can be selected to maintain genetic diversity within the breed. It is desirable that the breeding pair be as heterozygous (different) as possible in the traits being compared.
- Population studies - monitoring diversity, heterozygosity, inbreeding
.
Sample collection
In case of genetic profile testing, we prefer blood samples. The veterinarian will take the sample and at the same time confirm the identity of the individual by reading the microchip. The analysis is very sensitive to the quality of the DNA. A buccal swab sample can be used, but it must be very well collected and stored in a breathable container so that it dries well.
.
Methodology
Genomia Laboratory uses the internationally recognized and FCI recommended ISAG (International Society for Animal Genetics) standard. In quality tests ISAG repeatedly achieves the highest quality grade. DNA profiling is accredited by the Czech Institute for Accreditation according to ISO 17025, so Genomia has the highest possible competence for DNA profiling of dogs.
There are two main technological approaches to genetic profiling: STR and SNP. These approaches are not compatible with each other and cannot be compared.
.
STR genetic profile (ISAG2006)
STR (Short Tandem Repeats) genetic profile is based on the analysis of microsatellite markers - short repetitive DNA sequences of two to seven base pairs. The number of these repeats is individual and inherited from parents, making STR profiles an excellent tool for parentage verification and genetic identification.
The STR analysis process involves several main steps:
- DNA isolation - DNA is extracted from the sample to serve as the starting material for analysis.
- Amplification by polymerase chain reaction (PCR) - Specific stretches of DNA containing STR markers are amplified by PCR. Each STR marker is specifically amplified using primers - short sequences of DNA, with one of the primers labeled with a fluorescent dye at the 5' end, allowing the product to be detected.
- Fragment separation by electrophoresis - Multiplied DNA fragments are placed in a capillary electrophoretic device where they are separated by length in an electric field. Each allele of the STR marker corresponds to a specific fragment length, which depends on the number of repeats of the sequence motif.
- Detection and data analysis - Fluorescently labelled DNA fragments are detected by a laser scanner. The result is an electrophoretic recording that shows the length of each STR marker.
- Interpretation of results - The result is a unique combination of markers that identifies the individual. For each marker, the individual inherits one value from the father and one from the mother. By comparing STR profiles, it is possible to verify the identity of an individual or confirm relationships with more than 99.99% confidence.
- List of markers included in the ISAG2006 panel: AHTk211, CXX279, REN169O18, INU055, REN54P11, INRA21, AHT137, REN169D01, AHTh260, AHTk253, INU005, INU030, Amelogenin, FH2848, AHT121, REN162C04, AHTh171, REN247M23, AHTH130, REN105L03, REN64E19. Genomia reports at least 19 markers.
.
SNP genetic profile (ISAG2020)
SNP (Single Nucleotide Polymorphisms) genetic profile focuses on the analysis of point polymorphisms, i.e. changes in the DNA sequence at the level of single nucleotides. These changes are very stable and are inherited according to Mendel's laws, ensuring high reliability and more accurate and robust genetic identification. SNP profiles can be well used for population studies because they contain many markers evenly distributed across all chromosomes. This ensures complete coverage and representative representation for the assessment of heterozygosity and diversity. SNP profiling uses modern sequencing technologies for massively parallel sequencing, which increases the reliability and efficiency of testing.
The analysis of the SNP takes place in several steps:
- DNA isolation - The emphasis is on high quality of the sample - the most suitable material is blood.
- Massively Parallel Sequencing (MPS) - The modern method of massively parallel sequencing, or next generation sequencing (NGS), is used to analyse SNPs, allowing hundreds to thousands of genetic variants to be read simultaneously.
- SNP marker identification - Each SNP marker represents a specific position in DNA where single nucleotide variation occurs. ISAG2020 panels 1 and 2 standardize the analysis of 231 SNP markers, allowing international comparison of results. Genomia reports a minimum of 218 markers.
- Bioinformatics analysis - After sequencing, the data is processed by bioinformatics software that determines the genotype of an individual based on the presence of specific SNP variants. Each SNP marker carries “two letters” (two nucleotide bases) in the result, each of which the individual inherited from a different parent.
Summary of the benefits of SNP profiling:
- Higher accuracy - tracking more markers evenly distributed across all chromosomes allows for more accurate identification of individuals and relationships.
- Higher marker stability - one mutation occurs on average every 100 million bases per generation (in comparison, an STR marker has a mutation occurring on average every 1,000-10,000 transmissions of that marker)
- Higher throughput of the technological method - analysis of hundreds of samples in one time
- More suitable for population studies - monitoring inbreeding, genetic diversity, homozygosity/heterozygosity. Genomics reports the heterozygosity value for each SNP profile.
The disadvantages are longer sample processing time and higher costs for analysis and necessary technologies (powerful computers, specialized software for results evaluation, large data repositories, MPS sequencer validation). On the other hand, due to the high performance of the technology, the cost of analysis decreases with the number of samples tested.
.
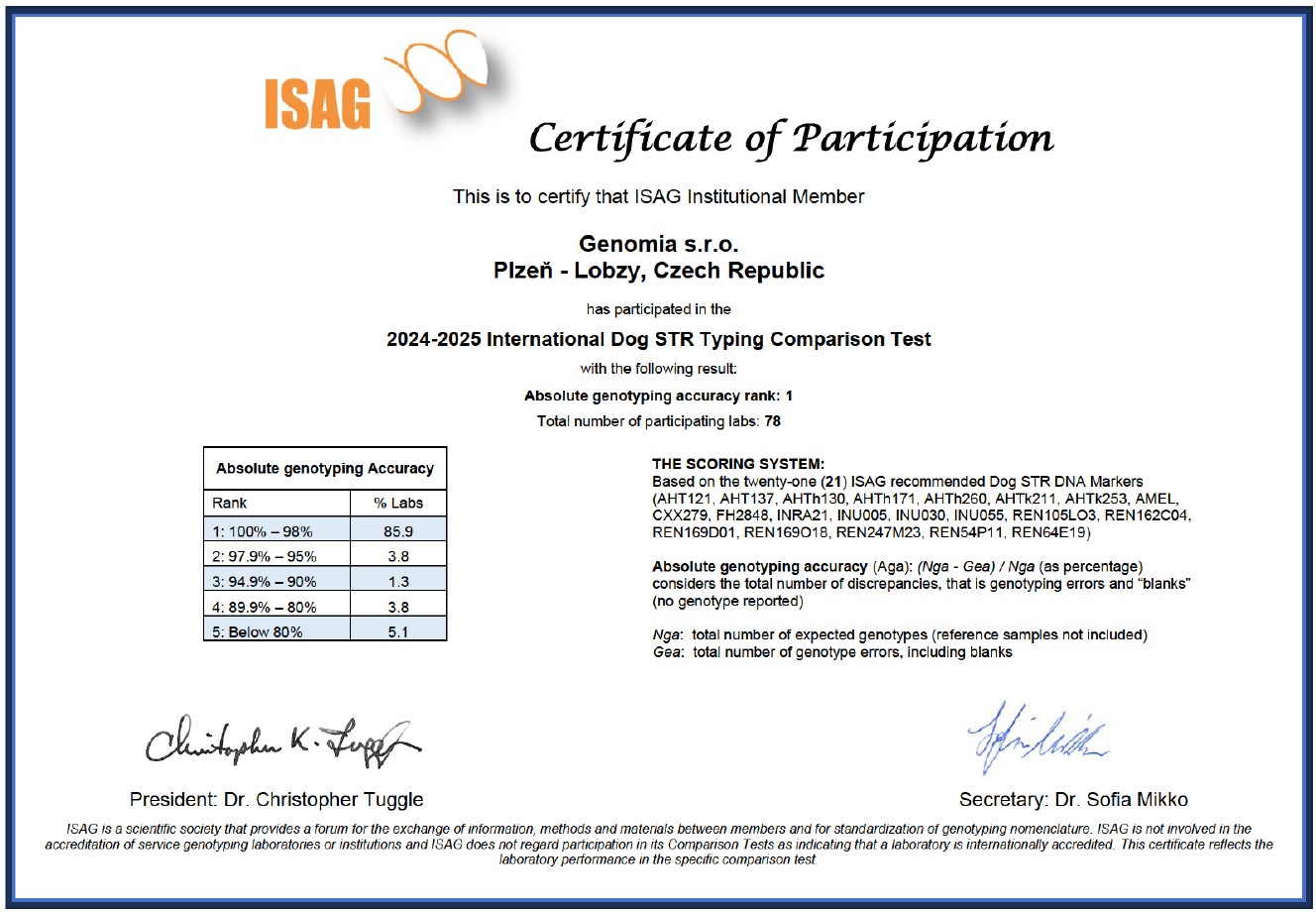
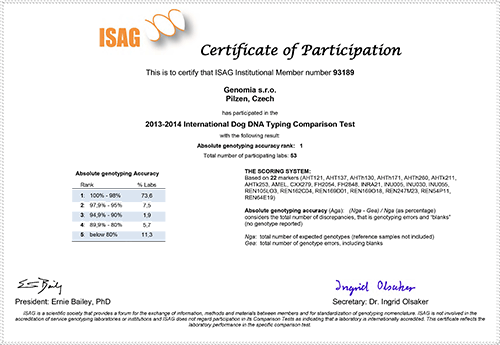
Genomia is an ISAG member
Genomia regularly participates in comparative quality tests, that are every two years held by ISAG (International Society for Animal Genetics). Here, you can see our quality certificates - Genomia has always succeed in quality rank 1:-)
.
.
.
.
.
.
.